spots in yeasts
A Napari plugin segmenting yeast cells and fluo spots to extract statistics.
A Napari plugin segmenting yeast cells and fluo spots to extract statistics.
The skeleton on this napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
Introduction
This Napari plugin's purpose is to extract statistics about fluo spots in yeast cells. We produce a segmentation of cells (based on the brightfield) and a segmentation of spots (based on the fluo channel). Then, we associate the measures to each cells.
Unless you use the NapariJ plugin to open images, or the cast_extension.ijm script to cast files, you can only launch this plugin on .tif images.
For now, the code produces JSON files compiling the metrics such as:
- The number of spots per cell.
- The average intensity of a spot.
- The area of each spot.
- The location of each spot.
We provide cast_extension.ijm which is another script meant to be used in Fiji/ImageJ. It is able to convert .nd and .czi images into basic .tif images so you can open them in Napari.
You can process your images either in one-shot (image per image) or in batch mode (by providing the path to a folder). In case you used batch mode, a control image is created so you can quickly check whether your segmentation was correctly performed.
Required packages in your environment:
naparicellposenumpyskimagetermcolormatplotlibcv2
Installation
You can install spots-in-yeasts via pip:
pip install spots-in-yeasts
To install latest development version :
pip install git+https://github.com/MontpellierRessourcesImagerie/spots-in-yeasts.git
Example
- Your images must have exactly two channels. The number of slices in each channel is totally up to you.
- First channel: fluo spots, second channel: brightfield.
- If you want to use the batch mode, you must use
.tifimages.
The two following images are the brightfield and fluo spots channels of the same image:
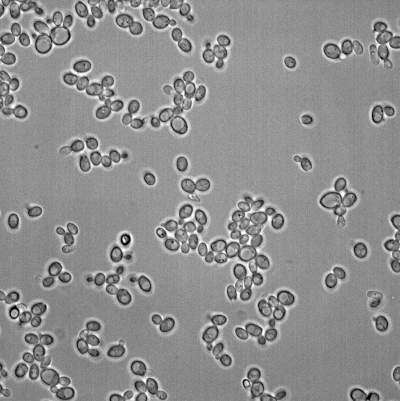
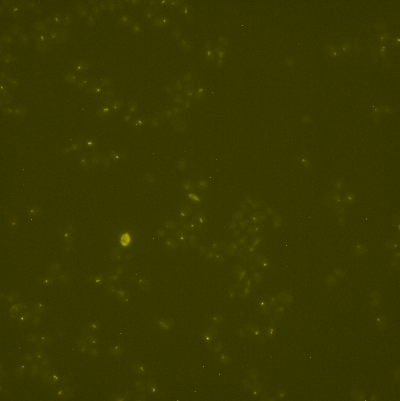
The following images are the cells labels and the spots positions:
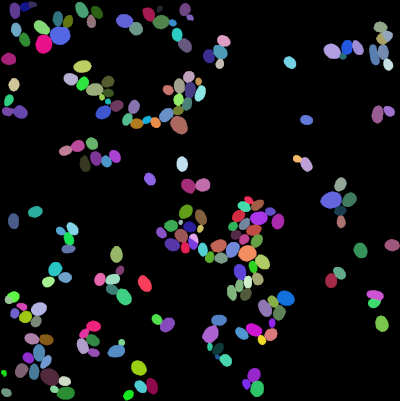

Usage
One-shot
- Open Napari. Keep the terminal opened, it provides lots of information.
- Before starting, make sure that no layer is currently open. You can clear your viewer with the
Clear layersbutton. - Drag'n'drop your image into the Napari viewer. It should show up in the left column.
- Click the
Split channelsbutton to separate the brightfield and the fluo on two different layers. Now, you should have two layers named "brightfield" and "fluo-spots". - To segment yeast cells, click the
Segment cellsbutton. The interface will certainly freeze during a few seconds (~10/30s). A new layer should appear, containing a value of intensity for each individual cell. - Click on the
Segment spotsbutton. This is a pretty fast operation. A new layer containing spots just appeared. Spots are represented as small white dots. You can change that in the layer's settings you struggle controling the result. - Finally, you can use the
Extract statsbutton to create a JSON file. This file will automatically be opened in your default text editor, but it is a temporary file, which means that it is not saved anywhere and will get lost if you don't save it yourself. - Once you are done, you can press the
Clear layersbutton again and pass to your next image, repeating the previous steps.
Batch mode
- Before starting, you need a folder containing correctly formated
.tiffiles. - Open Napari, and keep the terminal opened to see provided information.
- Set the
input folderfield to your folder containing.tifimages. - Set the
output folderfield to the path of a folder (preferably empty) that will receive the control images and the JSON files generated by the script. - You can click the
Run batchbutton to launch the process.
Note: In batch mode, your viewer won't show anything. You must rely on the terminal's content and the progress bar to know what is going on. To open the progress bar in Napari, click on activity in the lower-right corner.
Messages:
Export directory set to: /some/path/to/output: Folder provided by the user to receive produced files (JSON, controls)===== Working on: d1-230421-11S_2 (1/32)====: Name of the image currently processed and its rank. For example here, "d1-230421-11S_2" is being processed and it is the first image processed from a folder containing 32 images.Selected slices: (4, 8). (11 slices available): The script doesn't use all the slices in the image. Instead, it detects the most is-focus slice and takes N slices before and after it. In this example, 11 slices were available in the image. We are using the slices 4, 5, 6, 7, 8 for processing, so the most in-focus one is the 6th.Segmenting cells...: Notification that the script started segmenting yeasts cells.Cells segmentation done. 219 cells found.: End of cells segmentation. This message also provides you with the number of indiviual detected. This number is displayed before labels touching the borders are removed.Segmented cells from d1-230421-11S_2 in 10.0s.: Operations are timed. This is just the time report of cells segmentation.Starting spots segmentation...: Notification that the script started segmenting spots in the fluo channel.102 spots found .: Number of spots detected during the segmentation.Segmented spots from d1-230421-11S_2 in 1.0s.: Duration elapsed during spots segmentation.Spots exported to: /some/path/to/output/d1-230421-11S_2.json: Path to the exported metrics.Focused slice too far from center!: We don't use all the slices available. We detect the most in-focus one and take N slices before and after. This message means that there isn't N slices available after (or before) the most in-focus one. The process won't get interupted, but you want to be more careful about the segmentation of this image.The image d1-230421 BG- failed to be processed.: A basic sanity check is applied to the results before they get exported to reduce the amount of manual checking to perform. This message simply means that either the cells segmentation, or the spots segmentation is so bad that this image will be skipped.========= DONE. (288.0s) =========: Indicates that all the images contained in your folder were processed, the batch is over. The total amount of time if also displayed.
Contributing
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
License
Distributed under the terms of the MIT license, "spots-in-yeasts" is free and open source software
Issues
If you encounter any problems, please file an issue along with a detailed description.
Version:
- 1.2.0
Last updated:
- 2023-08-30
First released:
- 2023-08-29
License:
- MIT
Supported data:
- Information not submitted
Open extension:
Save extension:
Operating system:
- Information not submitted
Requirements:
- numpy
- magicgui
- magic-class
- qtpy
- opencv-python
- matplotlib
- termcolor
- scikit-image
- tifffile
- cellpose
- napari
- tox ; extra == 'testing'
- pytest ; extra == 'testing'
- pytest-cov ; extra == 'testing'
- pytest-qt ; extra == 'testing'
- napari ; extra == 'testing'
- pyqt5 ; extra == 'testing'





