napari-yolov5
Plugin adapted from Ultralytics to bring YOLOv5 into Napari
Plugin adapted from Ultralytics to bring YOLOv5 into Napari.
Training and detection can be done using the GUI. Training dataset must be prepared prior to using this plugin. Further development will allow users to use Napari to prepare the dataset. Follow instructions stated on Ultralytics Github to prepare the dataset.
The plugin includes 3 pre-trained networks that are able to identify mitosis stages or apoptosis on soSPIM images. More details can be found on the pre-print.
This napari plugin was generated with Cookiecutter using @napari's cookiecutter-napari-plugin template.
Installation¶
First install conda and create an environment for the plugin
conda create --prefix env-napari-yolov5 python=3.9
conda activate env-napari-yolov5You can install napari-yolov5 and napari via pip:
pip install napari-yolov5
pip install napari[all]For GPU support :
pip uninstall torch
pip install torchvision==0.10.0+cu111 -f https://download.pytorch.org/whl/torch_stable.htmlUsage¶
First select if you would like to train a new network or detect objects.

For Training :
Data preparation should be done following Ultralytics' instructions.
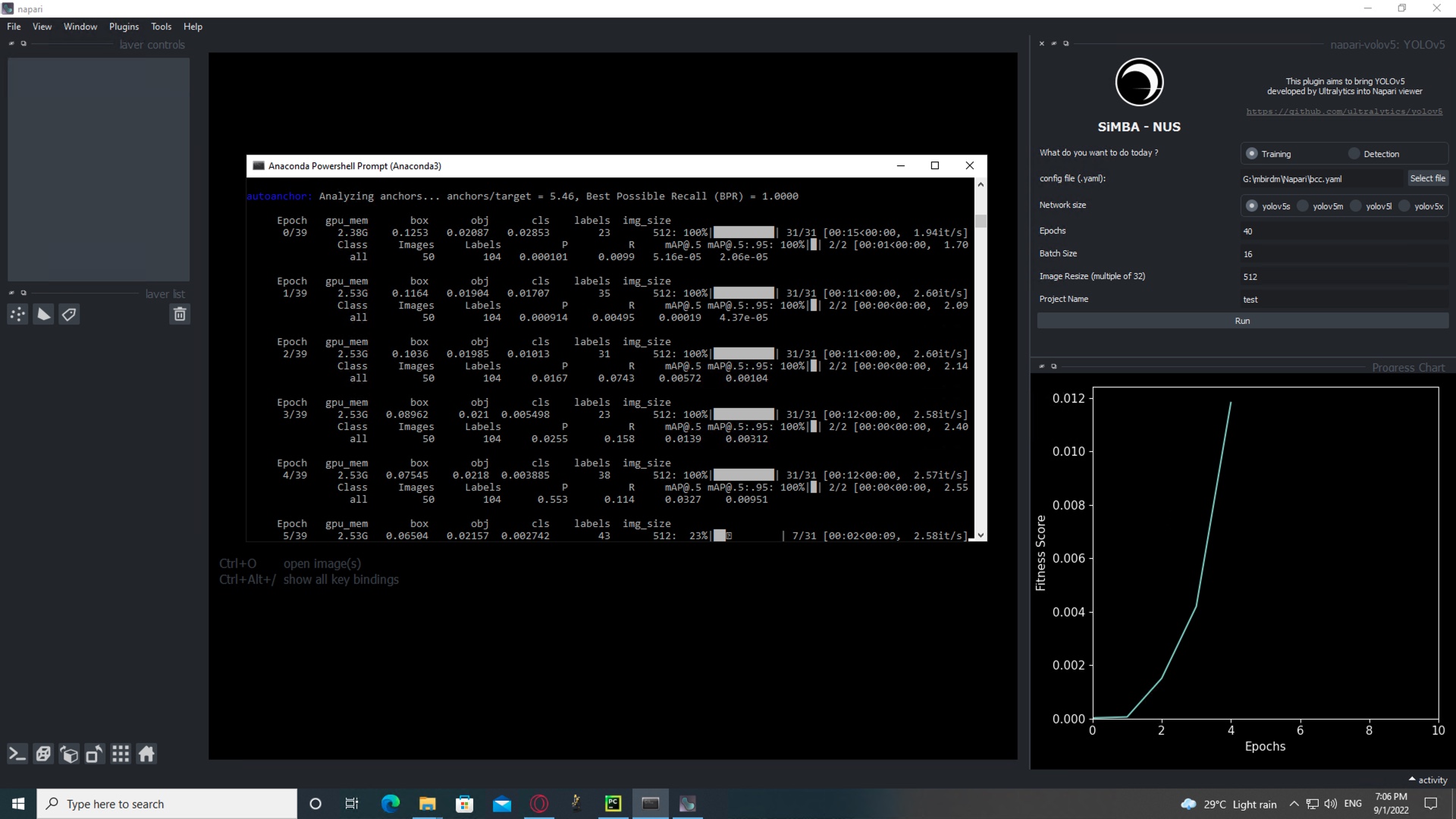
Select the size of the network, the number of epochs, the number of images per batch to load on the GPU, the size of the images (must be a stride of 32), and the name of the network.

An example of the YAML config file is provided in src/napari_yolov5/resources folder.
Progress can be seen on the Terminal or on the right-hand side of the viewer.
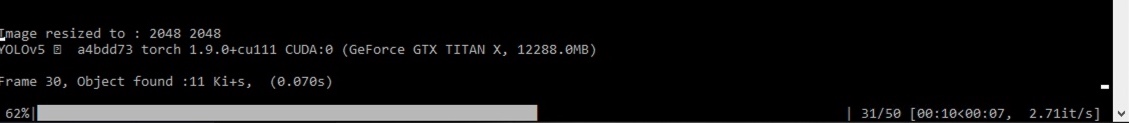
For Detection :
It is possible to perform the detection on a single layer chosen in the list, all the layers opened, or by giving a folder path. For folder detection, all the images will be loaded as a single stack.

Nucleus size of the prediction layer has te be filled to resize the image to the training dataset. Nucleus size of the training dataset will be asked in case of a custom network.
Confidence threshold defines the minimum value for a detected object to be considered positive. iou nms threshold (intersection-over-union non-max-suppression) defines the overlapping area of two boxes as a single object. Only the box with the maximum confidence is kept. Progress can be seen on the Terminal.
Few options allow for modification on how the boxes are being displayed (default : box + class + confidence score ; box + class ; box only) and if the box coordinates and the image overlay will be exported. Post-processing option will perform a simple 3D assignment based on 3D connected component analysis. A median filter (1x1x3 XYZ) is applied prior to the assignment. The centroid of each object is then saved into a new point layer as a 3D point with a random color for each class.
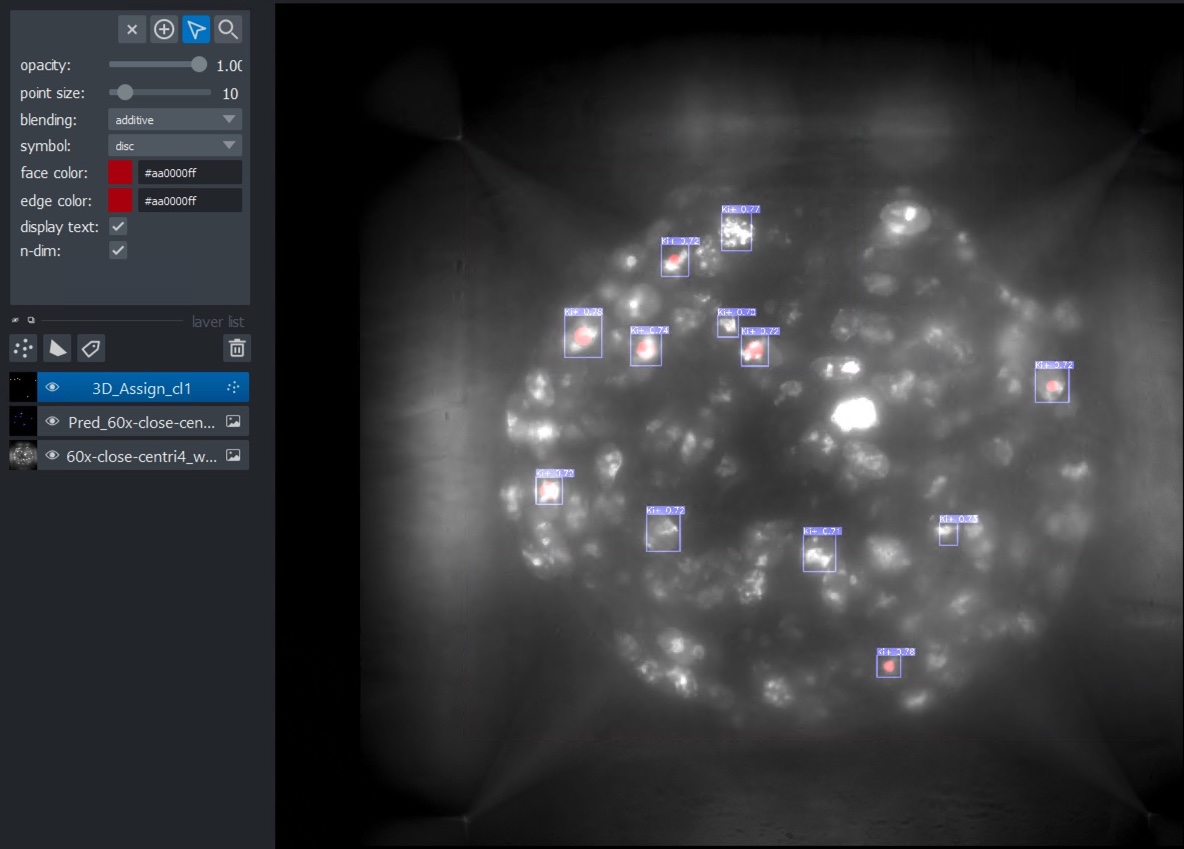
The localisation of each centroid is saved and the path is shown in the Terminal at the end of the detection. It is also possible now to define the export folder.
Contributing¶
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
License¶
Distributed under the terms of the GNU GPL v3.0 license, "napari-yolov5" is free and open source software
Issues¶
If you encounter any problems, please [file an issue] along with a detailed description.
Supported data:
- Information not submitted
Plugin type:
GitHub activity:
- Stars: 7
- Forks: 1
- Issues + PRs: 3



